GBR - DNA analysis to better understand thermal tolerance in corals (AIMS)
This study forms part of MTSRF Project 2.5i.2c and examines variations in coding regions (genes) of coral DNA and their relative frequency as distributed along a thermal gradient on the Great Barrier Reef. The genes were also investigated for their known or possible roles as an indicator of environmental stress, with particular emphasis on measuring thermal tolerance.
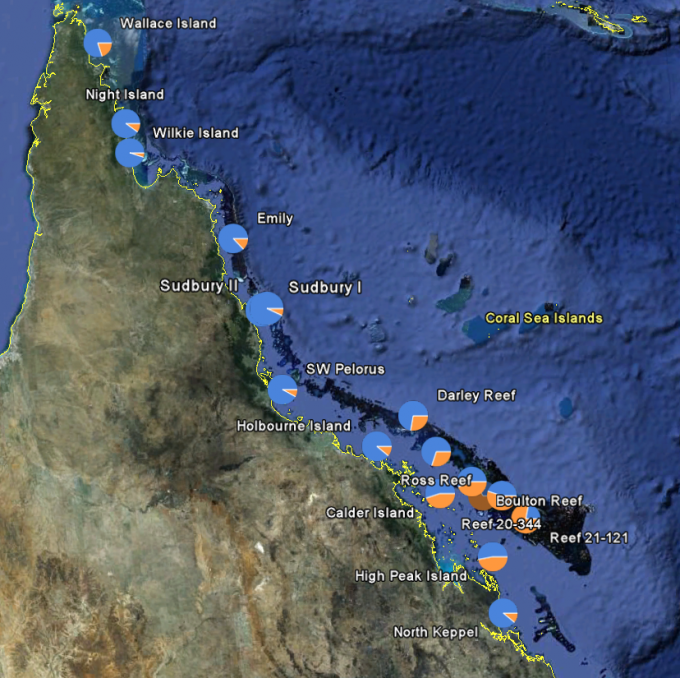
Samples were extracted from colonies at the 17 locations on the map. As a first step, transcribed DNA (mRNA) was extracted from several colonies at each of three thermally distinct populations along the GBR (Princess Charlotte Bay, Magnetic Island, Keppel Islands). RNA sequences were generated using next generation sequencing techniques (454 pyrosequencing). The outcome was over 500000 DNA sequences, containing over 80000 potential SNPs (Single Nucleotide Polymorphisms).
In a second step, 26 SNPs located in 16 genes (chosen for their known role in the oxidative stress response, or for the distribution of the SNPs in the original dataset) were investigated for their suitability to be included in a SNP genotyping assay. After assay optimization, 15 SNPs were chosen to be included in a large scale genotyping effort. Approximately 20 individuals from 17 reefs along the entire GBR locations have been genotyped at these 15 loci and the relative frequency of each nucleotide has been mapped at each site (use the e-atlas maps to view).